GWAS Meta-Analysis of Suicide Attempt: Identification of 12 Genome-Wide Significant Loci and Implication of Genetic Risks for Specific Health Factors
Abstract
Objective:
Suicidal behavior is heritable and is a major cause of death worldwide. Two large-scale genome-wide association studies (GWASs) recently discovered and cross-validated genome-wide significant (GWS) loci for suicide attempt (SA). The present study leveraged the genetic cohorts from both studies to conduct the largest GWAS meta-analysis of SA to date. Multi-ancestry and admixture-specific meta-analyses were conducted within groups of significant African, East Asian, and European ancestry admixtures.
Methods:
This study comprised 22 cohorts, including 43,871 SA cases and 915,025 ancestry-matched controls. Analytical methods across multi-ancestry and individual ancestry admixtures included inverse variance-weighted fixed-effects meta-analyses, followed by gene, gene-set, tissue-set, and drug-target enrichment, as well as summary-data-based Mendelian randomization with brain expression quantitative trait loci data, phenome-wide genetic correlation, and genetic causal proportion analyses.
Results:
Multi-ancestry and European ancestry admixture GWAS meta-analyses identified 12 risk loci at p values <5×10–8. These loci were mostly intergenic and implicated DRD2, SLC6A9, FURIN, NLGN1, SOX5, PDE4B, and CACNG2. The multi-ancestry SNP-based heritability estimate of SA was 5.7% on the liability scale (SE=0.003, p=5.7×10–80). Significant brain tissue gene expression and drug set enrichment were observed. There was shared genetic variation of SA with attention deficit hyperactivity disorder, smoking, and risk tolerance after conditioning SA on both major depressive disorder and posttraumatic stress disorder. Genetic causal proportion analyses implicated shared genetic risk for specific health factors.
Conclusions:
This multi-ancestry analysis of suicide attempt identified several loci contributing to risk and establishes significant shared genetic covariation with clinical phenotypes. These findings provide insight into genetic factors associated with suicide attempt across ancestry admixture populations, in veteran and civilian populations, and in attempt versus death.
Suicide accounted for more than 700,000 deaths worldwide in 2019 and was the fourth leading cause of death among 15- to 29-year-olds (1). Suicide attempt is even more common (2–4). Suicide attempt is strongly associated with psychiatric conditions, poor quality of life, traumatic experiences, and social and economic burden (1) and is the single strongest predictor of future suicide death (5).
Heritability estimates for suicidal thoughts and behaviors from twin and family studies range from 30% to 55% (6), and recent large-scale genome-wide association studies (GWASs) have yielded promising and replicable results. The International Suicide Genetics Consortium (ISGC) (total N=549,743; 29,782 cases) identified two loci reaching genome-wide significance for suicide attempt in individuals of primarily European ancestry admixtures, on chromosomes 6 (index SNP: rs71557378; p=1.97×10−8) and 7 (index SNP: rs62474683; p=1.91×10−10) (7). The intergenic locus on chromosome 7 remained significant after conditioning on psychiatric disorders and was independently replicated (p=3.27×10−3) (8) in the Million Veteran Program (MVP) cohort (9). The MVP cohort GWAS of suicide attempt (total N=409,153; 14,089 cases) resulted in two genome-wide significant multi-ancestry loci, on chromosomes 20 (index SNP: rs56817213; p=3.64×10−9) and 1 (index SNP: rs72730526; p=3.69×10−8) (8). A top signal identified at the dopamine receptor D2 locus (p=1.77×10−7) also showed moderate association in the ISGC GWAS (p=7.97×10−4) (7).
These studies established the complexity of the common variant genetic architecture of suicide attempt and demonstrated the critical role of sample size for discovering novel, replicable risk loci for suicide phenotypes through GWASs (10). Together, these GWASs suggested that larger studies will identify additional genomic risk loci and refine genetic risk metrics.
The objective of the present study was to conduct a meta-analysis of the ISGC and MVP studies (total N=958,896; 43,871 suicide attempt and suicide death cases). Moreover, there is considerable need to increase the diversity and generalizability of GWAS data (11). Combining all ISGC and MVP cohorts allowed for the largest GWAS meta-analyses of European, African, and East Asian ancestry admixtures to date. We also tested for gene set enrichment and functional follow-up specific to all included ancestral admixture populations.
Methods
GWAS Cohorts and Phenotype Ascertainment
The International Suicide Genetics Consortium (ISGC) cohort.
The ISGC analyses included 29,782 cases of suicide attempt (SA; defined as self-injurious behaviors with an intent to die) and/or suicide death and 519,961 control subjects from 18 cohorts (15 of SAs, two of suicide deaths, and one of both), 12 of which were ascertained clinically for the purpose of studying psychiatric disorders. Details about the specific cohorts have been provided previously (7), and cohort references and ascertainment methods are summarized in Table S1 in the online supplement. Twelve SA cohorts ascertained information on SA via in-person structured psychiatric interviews conducted by trained clinicians/researchers, two SA cohorts used self-report, and two SA cohorts used ICD codes or hospital records. All interviews and self-report items asked explicitly about SA rather than self-harm (which would also include nonsuicidal self-injury). ICD codes were coupled with information from emergency department settings and information on reason for contact and attempt methods that were mined from physician notes, in order to maximize evidence that suicidal intent was present. For the cohorts in which interviews or self-report were used to ascertain SA information, the SA was nonfatal. In an additional two cohorts, cases of suicide death were explicitly ascertained. The majority of suicide death cases were ascertained from the Utah Office of the Medical Examiner (hereafter “Utah”; N=4,692). In these cases, suicide cause-of-death determination results from a detailed investigation of the scene of the death and circumstances of death, determination of medical conditions by full autopsy, review of medical and other public records concerning the case, interviews with survivors, and standard toxicology workups (12). Suicide determination is traditionally conservative given its impact on surviving relatives. In the 746 suicide deaths from Kobe, Japan, autopsies were performed and cause of death was determined through discussion with the Medical Examiner’s Office and the Division of Legal Medicine at the Kobe University Graduate School of Medicine. The Columbia University cohort of both SA and suicide death included 317 suicide deaths that were determined by psychological autopsy and the coroner or medical examiner. A psychological autopsy is a method of determining the psychological factors that may have contributed to a death, considering additional information from family members, friends, acquaintances, medical records, and other relevant documents to better characterize a death of uncertain cause, including suspected suicides.
The Million Veteran Program cohort.
MVP recruitment and study procedures have been described previously (8). Participants provided a blood sample, consented to genetic analyses and the linking of their genetic information to the VA’s electronic health record system (EHR), and completed two optional surveys (9, 13). SA was defined as an act of deliberate self-harm with the intent to cause death that occurred at any point over the lifetime. Briefly, cases were defined as veterans with a documented history of SA in the EHR (N=14,089) and controls were defined as veterans with no documented history of suicidal thoughts or behaviors in the EHR (N=395,064). VA EHR sources were utilized to create an SA phenotype using 1) diagnostic codes for intentional self-harm, 2) suicidal behavior reports from the VA’s Suicide Prevention Applications Network database, and 3) mental health survey responses from the VA’s Mental Health Assistant database indicating a history of attempting suicide. Veterans who had a history of suicidal ideation but no SA were excluded from analysis. For all ISGC and MVP cohorts, it remains undetermined which individuals with SA may have died later by suicide. Details of sample sizes by genetic ancestry admixture for the ISGC and MVP cohorts are presented in Table 1.
| Cohort | Suicide Attempt/Death | Ascertainment | Cases | Controls |
|---|---|---|---|---|
| EUR | | | | |
| Army STARRS | Attempt | Military | 670 | 10,637 |
| Australian Genetics of Depression Study | Attempt | Psychiatric | 2,792 | 20,193 |
| Columbia University | Attempt and death | Psychiatric | 577 | 1,233 |
| GISS | Attempt | Psychiatric | 660 | 660 |
| German Borderline Genomics Consortium | Attempt | Psychiatric | 481 | 1,653 |
| iPSYCH | Attempt | Population | 7,003 | 52,227 |
| Janssen | Attempt | Psychiatric | 255 | 1,684 |
| Million Veteran Program | Attempt | Military | 9,196 | 287,370 |
| PGC Bipolar Disorder | Attempt | Psychiatric | 3,214 | 17,642 |
| PGC Eating Disorders | Attempt | Psychiatric | 170 | 5,070 |
| PGC Major Depressive Disorder | Attempt | Psychiatric | 1,528 | 16,626 |
| PGC Schizophrenia | Attempt | Psychiatric | 1,640 | 7,112 |
| UK Biobank | Attempt | Population | 2,433 | 334,766 |
| University of Utah | Death | Population | 4,692 | 20,702 |
| Yale-Penn | Attempt | Psychiatric | 475 | 1,817 |
| Total | | | 35,786 | 779,392 |
| EAS | | | | |
| CONVERGE Consortium | Attempt | Psychiatric | 1,148 | 6,515 |
| Kobe University | Death | Population | 746 | 14,049 |
| Million Veteran Program | Attempt | Military | 115 | 4,082 |
| Total | | | 2,009 | 24,646 |
| AFR | | | | |
| Grady Trauma Project | Attempt | General medical | 669 | 4,473 |
| Million Veteran Program | Attempt | Military | 3,507 | 74,306 |
| Yale-Penn | Attempt | Psychiatric | 629 | 2,902 |
| Total | | | 4,805 | 81,681 |
| LAT | | | | |
| Million Veteran Program | Attempt | Military | 1,271 | 29,306 |
| Multi-ancestry total | | | 43,871 | 915,025 |
TABLE 1. Summary of GWAS cohorts and primary ancestry admixturesa
Genotyping, Quality Control, and Imputation
Details of genotyping, quality control (QC), and imputation for the ISGC and MVP data sets have been described previously (7, 8). In the ISGC analyses, genotyping was performed locally by each of the research teams, using comparable procedures (8) (details per cohort are available in Table S1 in the online supplement). Standard parameters were used to retain individuals and SNPs after quality control for missingness, relatedness, and Hardy-Weinberg equilibrium. Genetic ancestry was defined by the contributing cohorts, and all ascertainment, QC, and analysis details of the ISGC and MVP cohorts are provided in Table S1 in the online supplement. Imputation was performed using the largest available ancestrally matched reference panels, either from the 1000 Genomes Project or the Haplotype Reference Consortium. We confirmed the comparability of imputation across the cohorts by comparing the final set of SNPs in the meta-analysis, including the number of cohorts in which they were present, and the INFO scores across cohorts and within ancestral admixture groups. Sample overlap and/or cryptic relatedness across cohorts was assessed and corrected for using the meta-analytic tools described below. Eight of the cohorts had high control:case ratios (using an arbitrary cutoff of >15:1). In these cases, the linkage disequilibrium (LD) score regression (LDSC) (14) attenuation ratio statistics were examined for evidence of population stratification or uncontrolled type I error in the cohort. For any evidence of inflation, the intercept was used to adjust the standard error of the summary statistics.
GWAS Meta-Analysis of Suicide Attempt
For both the ISGC and MVP cohorts, the initial GWAS analysis was conducted within genetic ancestral admixtures. For the ISGC meta-analysis, GWASs were conducted within study and genetic ancestral admixtures, covarying for at least 10 principal components of genetic ancestry, genomic relatedness matrices, or factors capturing site of recruitment or genotyping batch, as required (7). For the MVP cohort, ancestry was assigned for four mutually exclusive ancestral groupings utilizing a previously defined approach harmonizing genetic ancestry admixtures and self-identified ancestry groupings (HARE) (15). Subsequent MVP GWAS analyses were performed within ancestral admixtures using PLINK2 (16), covarying for genetic ancestry principal components, age, and sex.
A multi-ancestry meta-analysis of SA GWAS summary statistics was conducted using an inverse variance-weighted fixed-effects model (standard error) in METAL (17), assuming shared risk effects across ancestry admixtures. SNPs with a mean weighted minor allele frequency of <1%, a mean weighted imputation INFO score <0.6, or SNPs present in <80% of the total effective sample size were removed to ensure adequate statistical power at every variant included. Ancestry admixture-specific GWAS meta-analyses were conducted with cohorts of significant European (EUR), African (AFR), and East Asian (EAS) ancestry admixtures using the same procedures. Only one primary ancestral admixture population, Hispanic/Latino (LAT), was limited to a single cohort and thus could not be meta-analyzed. Inflation of test statistics due to polygenicity or cryptic relatedness was assessed using the LDSC attenuation ratio ([LDSC intercept−1]/[mean of association chi-square statistics−1]). Resulting genome-wide significant (GWS) loci were defined as those with p<5×10−8 with LD r2>0.1, within a 3,000-kb window, based on the structure of the Haplotype Reference Consortium (HRC) EUR reference panel for the multi-ancestry meta-analysis, or the HRC ancestry-appropriate reference panel otherwise. GWS loci for SA were examined for heterogeneity across cohorts via the I2 inconsistency metric and forest plots.
Estimation of Heritability and Genetic Association With Other Disorders
LDSC (14) and covariate-adjusted LDSC (cov-LDSC) (18) methods were used to estimate the phenotypic variance in SA explained by common SNPs (SNP-based heritability, h2SNP) from the GWAS meta-analysis summary statistics. LD scores from the 1000 Genomes Project (EUR and EAS) were used to derive h2SNP for the multi-ancestry GWAS meta-analysis and meta-analyses of European and East Asian ancestry admixtures. To obtain acceptable attenuation ratios for Hispanic/Latino and African ancestry admixture h2SNP estimates, we used covariate-adjusted AMR LD scores (AMR referring to the “Ad Mixed American” super population code) from Pan UK Biobank (https://pan.ukbb.broadinstitute.org) and AA LD scores (AA referring to a U.S. population of African ancestry admixtures) from gnomAD v2.1.1 (19). h2SNP was calculated on the liability scale assuming a lifetime prevalence of 2% for SA in the general population (the middle of the range reported worldwide) (20). The default script of LDSC was used to exclude SNPs with minor allele frequency (MAF) <1% and INFO <0.9 and also to restrict variants to the list of approximately 1.2 million HAPMAP SNPs that are typically well imputed across data sets. h2 estimates remained stable across >2% and >5% MAF thresholds. The genetic correlation attributable to genome-wide SNPs (rG) was estimated between the ancestral admixture groups using the Popcorn package (21), and with a range of psychiatric disorders using LDSC and the largest available discovery GWAS meta-analysis summary statistics (22–33). The latter analyses were confined to European ancestry admixtures for consistency with the discovery summary data. Tests were Bonferroni corrected, adjusting for up to 18 phenotypes hypothesized to be associated with SA based on previous epidemiological association or previous evidence of genetic association in LD Hub (34). Previous LD Hub analyses in ISGC were pre-categorized manually into risk factor groups relevant to SA (5, 35, 36): autoimmune disease, neurologic disease, heart disease, hypertension, diabetes, kidney disease, cancer, alcohol use, smoking, pain, psychiatric, sleep, life stressors, socioeconomic, and education/cognition. Values for rG of SA in ISGC and MVP in this study were calculated using LDSC, and references for the discovery GWAS are listed in Table S2 in the online supplement. Differences in rG across other phenotypes using EUR GWAS meta-analyses were tested as a deviation from 0, using the block jackknife method implemented in LDSC (37). To examine phenome-wide partial genetic causality, the Complex-Traits Genetics Virtual Lab (CTG-VL) (38) was used to conduct false discovery rate–corrected genetic causal proportion (GCP) analyses on the EUR summary data.
Conditioning Suicide Attempt on Major Depressive Disorder and PTSD
The results of the EUR GWAS SA meta-analysis were conditioned on genetic risks for major depressive disorder (MDD) (27) and posttraumatic stress disorder (PTSD) (32) in secondary analyses, to examine genetic associations both shared with and unique to suicide risk. Results were conditioned because MDD and PTSD are both highly comorbid with SA, and because PTSD is particularly prevalent in military veteran populations (i.e., MVP). Conditioning was conducted using multitrait-based conditional and joint analysis using GWAS summary data (mtCOJO) (39), implemented in the GCTA software program (40). mtCOJO estimates the effect size of a SNP on an outcome trait (e.g., SA) conditioned on one or more exposure traits (e.g., MDD). GWS SNPs for the exposure are used as instruments to estimate the effect of the exposure on the outcome, and this effect is used to perform genome-wide conditioning, yielding conditioned effect sizes and p values for the outcome trait. The EUR-only SA GWAS summary statistics were used as the outcome trait, because mtCOJO requires GWAS summary statistics for the exposure trait, which were derived from EUR ancestry discovery GWAS. To select independent SNPs as instruments, we selected those that were more than 1 megabase apart or had an LD r2 <0.05 based on the 1000 Genomes Project Phase 3 EUR reference panel (41). mtCOJO is robust to sample overlap between the GWASs of the exposure and outcome. In this analysis, statistical power to detect genetic associations at individual SNPs was reduced relative to the unconditioned analysis by the additional model parameters, but the genetic correlations using the conditioned summary statistics provide valuable insights into the relevant risk factors for SA over and above those related to MDD and PTSD.
Gene, Gene Pathway, and Tissue Enrichment Analyses
Enrichment analyses of the GWAS results were performed to probe genes, biological pathways, and tissues implicated in SA, using the multi-ancestry and ancestry admixture-specific GWAS results. The p values quantifying the degree of association of genes and gene sets with SA were calculated using MAGMA v1.08 (42), implemented in FUMA v1.3.7 (43). Input SNPs were mapped to 18,627 protein-coding genes. Genome-wide significance was defined as a p value <2.68×10−6 (0.05/18,627). Curated gene sets that included at least 10 genes from MSigDB v7.0 were tested for association with SA. Competitive gene-set tests were conducted to correct for gene size, variant density, and LD within and between genes. Tissue-set enrichment analyses were also performed using MAGMA implemented in FUMA, to test for enrichment of association signal in genes expressed in 54 tissue types from GTEx v8 (44) (Bonferroni-corrected p threshold, 9.26×10−4).
Drug Target Enrichment Analyses
Additional gene-set enrichment analyses of both the multi-ancestry and EUR GWAS meta-analysis results were performed, restricted to genes targeted by drugs, in order to investigate putative relationships of suicide attempt with specific drug types. These analyses do not identify causal relationships but may implicate genes relevant to pharmacotherapy. This approach has been described previously (45). Gene-level and gene-set analyses were performed in MAGMA v1.08. Gene boundaries were defined using build 37 reference data from the National Center for Biotechnology Information, available on the MAGMA website (https://ctg.cncr.nl/software/magma), extended 35 kb upstream and 10 kb downstream to increase the likelihood of including regulatory regions outside of the transcribed region. Gene-level association statistics were defined as the aggregate of the mean and the lowest variant-level p value within the gene boundary, converted to a Z value. Gene sets were defined comprising the targets of each drug in the Drug-Gene Interaction Database (DGIdb) v.2 (46) and in the Psychoactive Drug Screening Ki Database (47), both downloaded in June 2016 (45). Analyses were performed using competitive gene-set analyses in MAGMA.
Results from the drug-set analysis were then grouped according to the Anatomical Therapeutic Chemical (ATC) class of the drug (45). Only drug classes containing at least 10 valid drug gene sets within them were analyzed, and drug-class analysis was performed using enrichment curves. All drug gene sets were ranked by their association in the drug-set analysis, and then for a given drug class, an enrichment curve was drawn scoring a “hit” if the drug gene set was within the class, or a “miss” if it was outside of the class. The area under the curve was calculated, and a p value for this was calculated as the Wilcoxon Mann-Whitney test comparing drug gene sets within the class to drug gene sets outside the class (45). Bonferroni-corrected significance thresholds of p<5.79×10−5 and p<4.35×10−4 were used for the drug-set and drug-class analyses, respectively, accounting for 863 drug sets and 115 drug classes.
Summary Data-Based Mendelian Randomization
Summary data-based Mendelian randomization (SMR) (v1.03) (48, 49) was applied to detect GWAS signals that colocalize with expression quantitative trait loci (eQTLs), in order to investigate putative causal relationships between SNPs and SA via gene expression. SMR was performed using eQTL summary statistics from the MetaBrain consortium (50), a cortex-derived eQTL data set consisting of 2,970 EUR cortex samples. The analysis was conducted using the EUR-only GWAS meta-analysis results, for consistency with the eQTL data. Brain eQTL data from comparable sample sizes in other ancestral groups are not currently available. SMR analysis was limited to transcripts with at least one significant cis-eQTL (p<5×10−8) in the data set (of 8,753 in MetaBrain). The Bonferroni-corrected significance threshold for the SMR analysis was p<5.71×10−6, and the significance threshold for the HEIDI test (heterogeneity in dependent instruments) (51) was p≥0.01. A nonsignificant HEIDI test suggests a direct causal role of the SA-associated SNPs on gene expression, rather than a pleiotropic effect.
Polygenic Risk Scoring
Polygenic risk scores (PRSs) for SA were tested for association with SA compared with controls in six target cohorts: Psychiatric Genomics Consortium Major Depressive Disorder, Bipolar Disorder, and Schizophrenia (all European ancestry admixtures); CONVERGE (East Asian ancestry admixtures); and Yale-Penn and Grady Trauma Project cohorts (both primarily African ancestry admixtures, located in the United States). The SA GWAS meta-analysis was repeated, excluding each cohort in turn, to create independent discovery data sets. PRSs were generated using PRS-CS (51), which uses a Bayesian regression framework to place continuous shrinkage priors on the effect sizes of SNPs in the PRSs, adaptive to the strength of their association signal in the discovery GWAS and the LD structure from an external reference panel. The 1000 Genomes EUR, EAS, or AFR reference panels (41) were used to estimate LD between SNPs, as appropriate for each target cohort. PLINK v1.9 (16) was used to weight SNPs by their effect sizes, calculated using PRS-CS, and to sum all SNPs into PRSs for each individual in the target cohorts. PRSs were tested for association with case-versus-control status in the target cohort using a logistic regression model including covariates as per the GWAS. The amount of phenotypic variance explained by the PRS (R2) was calculated on the liability scale, assuming a lifetime prevalence of 2% for SA in the general population (20). The Bonferroni-corrected significance threshold, adjusting for six tests, was p<0.008.
Results
Significant Shared Genetic Architecture of SA Between Civilian (ISGC) and Military (MVP) Populations
The multi-ancestry GWAS included 43,871 cases and 915,025 controls from 22 cohorts (Table 1). Cases were of predominantly European ancestry admixtures (EUR, 81%), with 11% of cases with significant African ancestry admixtures located in the United States (AFR), 5% with East Asian ancestry admixtures (EAS), and 3% with Hispanic/Latino ancestry admixtures located in the United States (LAT). Case definition was lifetime SA, with ∼13% of all cases having died by suicide. Additional information on study characteristics and ascertainment methods is presented in Table S1 in the online supplement.
Cohorts across ISGC and MVP differed with respect to ascertainment, with ISGC being largely civilian and MVP being military (Table S1A). However, examination of the genetic correlation of EUR GWAS meta-analyses for ISGC and MVP (rG=0.81, SE=0.091, p=2.85×10−19) indicated consistency of common-variant genetic architecture across these meta-analyses. Results from both fixed and meta-regression models were comparable in the multi-ancestry and EUR GWAS meta-analyses (all GWS effect size correlations >0.99), indicating that ancestry and cohort ascertainment were unlikely to confound observed genetic effects (Table S1B).
12 GWS Loci Identified by GWAS Meta-Analysis of SA Across and Within Ancestries
The multi-ancestry GWAS meta-analysis identified eight GWS loci (p<5×10−8) (Figure 1). The h2SNP of SA was significant at 5.7% (SE=0.003, p=5.70×10−80) on the liability scale assuming an SA population prevalence of 2%. The cov-LDSC intercept was 1.04 (SE=0.01, p=1.59×10−5), and the attenuation ratio was 0.13 (SE=0.03), indicating that the majority of inflation of GWAS test statistics is likely due to polygenicity (see Figure S1 in the online supplement).
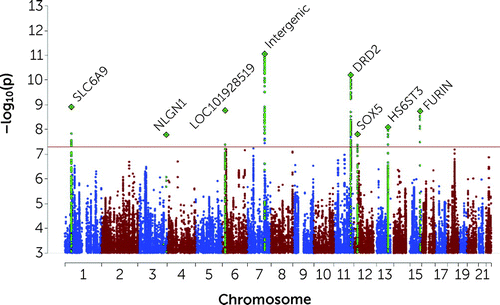
FIGURE 1. Manhattan plot of multi-ancestry GWAS meta-analysis of suicide attempta
aThe x-axis shows genomic position and the y-axis shows statistical significance as –log10(p). The horizontal line shows the genome-wide significance threshold (p<5.0×10−8). Labels represent the nearest gene to the index SNP. Regional plots of the eight genome-wide significant loci across ancestry admixture populations and the four genome-wide significant loci in subjects of European ancestry admixtures are presented in Figures S3–S14 in the online supplement.
The locus most strongly associated with SA was in an intergenic region on chromosome 7 (index SNP rs62474683 A allele, odds ratio=1.05, 95% CI=1.04–1.07, p=8.72×10−12; frequency in cases, 0.57; frequency in controls, 0.56; a forest plot is provided in Figure S2 in the online supplement). At other GWS loci, index SNPs were intronic in the SLC6A9, DRD2, HS6ST3, and FURIN genes (Table 2; see Table S1B in the online supplement for additional summary data on all GWS loci). On chromosome 3, a GWS SNP localized to the 5′ untranslated region of the NLGN1 gene, although the index SNP lacked neighboring SNPs in LD. There was no evidence of heterogeneity of effects across cohorts for any GWS locus according to I2 heterogeneity indices (see Table S1B in the online supplement). Forest plots for GWS loci are provided in Figures S2–S9 in the online supplement.
| CHR | Index SNP | BP | Locus Start..Stop | Nearest Gene (distance to index SNP in kb) | p | Odds Ratio | SE | A1 | A2 | Direction | NCohorts | NTotal | NEff |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Multi-ancestry | | | | | | | | | | | | | |
| 1 | rs3791129 | 44480093 | 44,462,155..44,497,134 | SLC6A9 (0.0) | 1.22E−09 | 1.055 | 0.009 | G | A | +-+++++--+-+?+++?+??++ | 18 | 933,136 | 158,078 |
| 3 | rs7649709 | 173129819 | 173,113,742..174,012,162 | NLGN1 (0.0) | 2.32E−08 | 1.054 | 0.010 | A | C | ++++++-+++-+?++-++++++ | 21 | 954,890 | 164,921 |
| 6 | rs62404522 | 19307114 | 19,068,774..19,180,711 | LOC101928519 (−76.4) | 1.68E−09 | 1.067 | 0.011 | C | T | +--+++--++++-++-+-++++ | 22 | 956,659 | 166,924 |
| 7 | rs62474683 | 115020725 | 114,763,653..114,871,409 | LINC01392 (−149.3) | 8.72E−12 | 1.054 | 0.008 | A | G | +-++++-++++++++-++-+++ | 22 | 958,896 | 167,455 |
| 11 | rs7131627 | 113299829 | 113,280,327..113,346,120 | DRD2 (0.0) | 6.2E−11 | 1.053 | 0.008 | G | A | +++++--+++++++++?-++++ | 21 | 944,101 | 164,620 |
| 12 | rs17485141 | 24213634 | 23,682,438..24,715,425 | SOX5 (0.0) | 1.54E−08 | 1.049 | 0.009 | C | T | +++-++++++++-+++++++++ | 22 | 958,896 | 167,455 |
| 13 | rs9525171 | 96908223 | 96,742,361..97,491,816 | HS6ST3 (0.0) | 8.07E−09 | 1.044 | 0.008 | C | G | +-+++++--+-+-+++++-+++ | 22 | 958,896 | 167,455 |
| 15 | rs17514846 | 91416550 | 91,411,818..91,426,687 | FURIN (0.0) | 1.81E−09 | 1.048 | 0.008 | C | A | ++++++++++-+-+++?+?+++ | 20 | 938,959 | 162,145 |
| EUR | | | | | | | | | | | | | |
| 1 | rs2503185 | 66461401 | 66,258,193..66,840,262 | PDE4B (0.0) | 3.42E−08 | 1.047 | 0.008 | A | G | ++++++---+++-++ | 15 | 815,178 | 136,860 |
| 6 | rs35869525 | 26946687 | 29,640,168..30,152,231 | (MHC) | 2.18E−08 | 1.089 | 0.015 | C | T | +?-+-+++-++++++ | 14 | 803,626 | 134,461 |
| 14 | rs850261 | 57346423 | 57,278,724..57,398,026 | OTX2-AS1 (0.0) | 1.37E−08 | 1.049 | 0.008 | A | G | ++-+-++++++++++ | 15 | 815,178 | 136,860 |
| 22 | rs2284000 | 37053338 | 36,956,904..37,099,797 | CACNG2 (0.0) | 1.98E−08 | 1.055 | 0.010 | C | G | ++++++-+-+-+?++ | 14 | 812,886 | 135,108 |
TABLE 2. Results from meta-analyses of suicide attempt showing the index SNP from each genome-wide significant locusa
The EUR GWAS meta-analysis h2SNP was estimated at 7.0% (SE=0.4%) and identified four additional GWS loci (Table 2; see also Figure S10 and forest plots in Figures S11–14 in the online supplement), composed of mostly intergenic index SNPs. The nearest genes were PDE4B, OTX2-AS1, CACNG2, and one locus was in the major histocompatibility complex. GWAS meta-analyses in AFR (h2SNP=9.8%, SE=1.8%) and EAS (h2SNP=9.8%, SE=4.5%) produced no GWS loci. The LAT SA h2SNP (from the MVP GWAS) was estimated at 10.0% (SE=6.5%). Regional plots of the 12 GWS risk loci across all meta-analyses are presented in Figures S15–S26 in the online supplement. Mapped genes from the top loci in multi-ancestry and ancestry admixture-specific meta-analyses are presented in Tables S3–S6 in the online supplement. Summary statistics from these GWASs are available through the Psychiatric Genomics Consortium data access portal.
Genetic Correlations of SA Across Ancestry GWASs
The genetic correlations of SA across each of the ancestral groupings were attenuated, with estimated rG values between 0.064 (SE=0.574) (EAS with LAT) and 0.997 (SE=0.537) (EUR with LAT); Popcorn rG results are provided in Table S7 in the online supplement. Individual cohort GWASs were variably powered to estimate genetic correlation estimates with the other cohorts. LDSC estimates across all individual GWASs are presented in Table S8 in the online supplement, although cov-LDSC h2SNP and Popcorn rG values in Table S7 in the online supplement are the preferred sources for statistics involving ancestry admixtures.
Enrichment of SA GWS Loci for Brain-Expressed Genes and Overlap With Previous Genetic Associations to Known Risk Factors
Significant signal enrichment was observed in genes expressed in pituitary gland and brain tissues, based on the multi-ancestry GWASs (see Table S9 in the online supplement). Significant gene expression in brain was also observed in the EUR analysis (see Table S10 in the online supplement). Tissue-set enrichment analyses and corresponding GTEx gene expression heat maps for all of the multi- and ancestry admixture-specific GWASs are provided in Tables S9–S12 and Figures S31–S34 in the online supplement.
Several GWS genes were identified in MAGMA analyses of the multi-ancestry and EUR meta-analyses (see Table S13 in the online supplement; enrichment of SA signal with genes and gene sets across all meta-analyses are presented in Tables S13–S14 in the online supplement). MAGMA gene-based tests of the GWAS meta-analyses, with GWS results, are presented in Manhattan plots and QQ plots in Figures S27–S30 in the online supplement. EAS and AFR p-value thresholds for inclusion of GWAS variants in follow-up analysis were relaxed to p<1×10−5 and 1×10−6, respectively, in order to explore gene-based tests of top ancestry-specific GWAS variants. Top genes implicated in the EAS analysis included C11orf87, MYO1C, and FAXC, and top genes implicated in the AFR analysis included CNTNAP2, IGF2R, MAN1B1, and SLC22A1. Neither set of genes was significantly associated with any pathway or tissue enrichment.
Gene-set analyses from the multi-ancestry and EUR GWAS identified 519 significant gene sets (31 and 488, respectively), spanning multiple domains, including epigenetics, gene regulation and transcription, cellular response to stress, DNA repair, and immunologic signatures (see Table S14 in the online supplement). The 31 multi-ancestry gene sets included schizophrenia and autism, containing protein-coding genes such as FURIN, FES, and DRD2, mapped from GWS loci. Most of the 488 EUR gene sets were due to overlap with a small group of 35 histone-coding genes.
Significant proportions of overlapping genes in GWAS Catalog (52) gene sets were observed for both multi-ancestry and EUR meta-analyses (see Figures S35–S36 in the online supplement). The 12 GWS loci from the multi-ancestry and EUR GWAS meta-analyses were tagged in several GWASs including cognition, smoking, insomnia, and risky behavior. Six of the 12 risk loci had p values <0.005 for the “Suicide or Other Intentional Self-Harm” analysis in FinnGen. A comprehensive list of results of SNP associations from the GWAS Catalog is presented in Table S15 in the online supplement. Examination of the phenome-wide association study results (p<0.005) across UK Biobank, FinnGen, and the GWAS Catalog resulted in the identification of several psychiatric, weight/BMI–related, and immune-related traits (see Table S16 in the online supplement).
Two loci implicated specific genes, FES and TIAF1, that were significantly associated with SA in SMR analyses and passed the HEIDI test. SMR results suggested that SA risk may be mediated by an increased expression of FES (previously implicated in cross-ancestry schizophrenia [53]) and decreased expression of TIAF1 in cortex (see Table S17 in the online supplement).
Significant Overlap of SA GWS Loci and Targets of Antipsychotics and Antidepressants
Drug target enrichment results suggested that SA risk is most associated with the targets of antipsychotic and antidepressant drug classes. In the multi-ancestry gene-set analysis of the targets of drug classes defined by their ATC classes (45), there was significant enrichment in the targets of four drug classes: Antipsychotics and Psychoanaleptics, which includes individually significant Antidepressants and its subclass Other Antidepressants (see Table S18 in the online supplement). The class Other Antidepressants includes those not classified as selective serotonin reuptake inhibitors, monoamine oxidase inhibitors, or monoamine reuptake inhibitors.
In the EUR ancestry admixture GWAS analysis, there was significant enrichment in the targets of just three drug classes, including Antipsychotics, the broad class of Psycholeptics (drugs with a calming effect on behavior), and the class Cytotoxic Antibiotics and Related Substances (see Table S19 in the online supplement). Only one drug, the insecticide cyfluthrin, was significantly enriched when grouping genes targeted by individual drugs (from DGIdb v.2 and the Psychoactive Drug Screening Ki Database), and this was observed only in the EUR GWAS results (see Tables S20 and S21 in the online supplement for multi-ancestry and EUR results).
Significant Genetic Correlation of SA With Known Nonpsychiatric Risk Factors Minimally Affected After Conditioning on MDD and PTSD
The out-of-sample polygenic risk analyses based on the new ISGC + MVP discovery GWAS meta-analysis statistics resulted in higher R2 estimates than were observed in previous ISGC analyses, particularly for the AFR cohorts, where the maximum variance explained (R2) was 0.66% (p=0.01), with a maximum increase of 146%, and EAS cohorts, where R2 was 0.34% (p=8.1×10−6), with a 36% increase. EUR maximum variance explained was 1.11% (p=6.2×10−22), a 24% increase from previous ISGC analyses (see Table S22 in the online supplement). Figure 2 presents a forest plot of the genetic correlations of the EUR GWAS meta-analyses of suicide attempt with several physical and mental health phenotypes, as well as one control phenotype (BMI). Significant shared genetic covariation of EUR SA with smoking (rG=0.46, SE=0.03, p=8.06×10−63), attention deficit hyperactivity disorder (ADHD) (rG=0.55, SE=0.04, p=2.98×10−41), risk tolerance (rG=0.32, SE=0.02, p=1.34×10−59), and chronic pain (rG=0.45, SE=0.03, p=9.50×10−50) were observed both before and after conditioning on genetic risks for MDD and PTSD. Significant positive genetic correlations of neuroticism, schizophrenia, bipolar disorder, and self-harm ideation with SA (rG=0.45, SE=0.03, p=1.0×10−52; rG=0.43, SE=0.03, p=1.32×10−55; rG=0.48, SE=0.04, p=1.81×10−37; rG=0.83, SE=0.06, p=1.94×10−51) did not remain significant after conditioning on both MDD and PTSD.
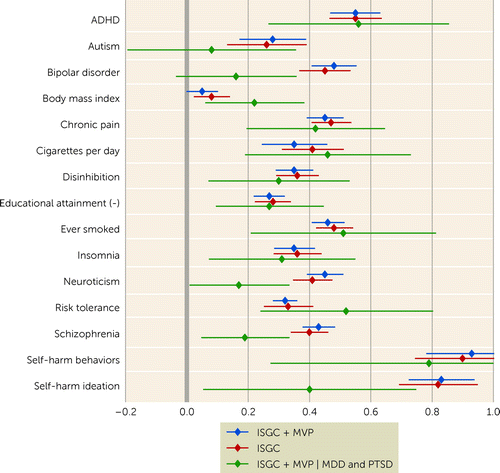
FIGURE 2. Forest plot of genetic correlations of the multi-ancestry GWAS meta-analyses of suicide attempt with physical and mental health phenotypesa
aThe x-axis presents genetic correlation values with 95% confidence intervals, and the y-axis presents the discovery GWAS for multiple phenotypes. ISGC=International Suicide Genetics Consortium meta-analysis; ISGC + MVP=the primary meta-analysis including GWASs from both ISGC and Million Veteran Program sets of cohorts; ISGC + MVP | MDD and PTSD=the combined GWAS meta-analysis of both cohorts conditioning on major depressive disorder and posttraumatic stress disorder.
For completeness of comparison across cohorts and phenotypic subgroups (SA versus suicide death), genetic correlation estimates for phenotypes are presented in Table S23 in the online supplement using the European ancestry admixture GWAS summary statistics from 1) ISGC + MVP, 2) ISGC only, 3) MVP only, 4) ISGC without suicide death, 5) ISGC suicide death only (the Utah Suicide Study; current N=4,692 EUR suicide deaths and 20,702 controls), and 6) conditioning on MDD and PTSD for MVP, ISGC, and MVP + ISGC. LDSC jackknife tests of differences between these genetic correlation estimates are presented in Table S24 in the online supplement, and more exhaustive comparisons of phenome-wide rG and genetic causal proportion analyses, with the European admixture GWAS meta-analysis, are provided in Table S25 in the online supplement. Genetic causal proportion analyses implicated several nonpsychiatric genetic risks in EUR SA, including particulate air matter pollution exposure (PM2.5), smoking exposures, and pulmonary health factors. Risk factors with significant partial genetic causality estimates are presented in Table S25.
Discussion
This study reflects the largest GWAS meta-analysis of SA to date, incorporating multiple ancestral admixture populations and expanding the set of GWS loci from four to 12. Discovery of three of the novel GWS loci, and improved out-of-sample PRS prediction across ancestry, was only possible with the aggregation of all ancestral admixture cohorts. The results show, for the first time, that implicated genes are highly expressed in brain tissue, are enriched in pathways related to gene regulation and transcription, cellular response to stress, DNA repair, and immunologic signatures, and are shared with epidemiological risk factors. Genetic correlation and causal proportion analyses implicate a number of nonpsychiatric genetic risks in SA, including pulmonary health factors. We also provide important evidence that a significant proportion of the common variant genetic architecture of SA is shared across large civilian and veteran populations with disparate demographic characteristics.
One advantage of combining the ISGC with MVP was the opportunity to examine genetic effects across heterogeneous cohorts. For example, the sample composition and ascertainment across the ISGC is predominantly civilian and international, with a larger proportion of females (7). A number of the ISGC samples from the Psychiatric Genomics Consortium cohorts (Table 1) are collected from individuals with major psychiatric disorders, representing a more clinical population. In contrast, the MVP cohorts are predominantly male (8), and all are military veterans ascertained through the U.S. Department of Veterans Affairs health care system. The consistency of SA common variant genetic architecture across EUR MVP and ISGC cohorts indicates that power may be further enhanced by combining future cohorts with differing ascertainments.
As expected, the increase in sample size, and the resulting increase in statistical power, led to the identification of several new GWS loci and improved out-of-sample PRS prediction, across ancestries, relative to the previous ISGC-only analyses. The loci identified in this study implicate genes expressed in brain. Genes associated with SA in this study are highly enriched among psychiatric phenotypes and overall health and wellness risk factors for SA. Brain is the predominant tissue enriched for associated genes, and there is also significant enrichment in pituitary gland, consistent with previous association of SA with hypothalamic-pituitary-adrenal system dysregulation (54). In addition, the enrichment of pathways related to epigenetics and gene regulation and transcription suggest that epigenetic modifications, such as DNA methylation, may play a role in modulating the effect of SA-associated genetic variants. However, epigenetic pathways were only enriched in GWASs of European ancestry admixtures, pointing to the potential importance and varied impact of epigenetic mechanisms in diverse biological systems that may contribute to SA risk. Pathways enriched in the multi-ancestry GWAS were absent of histone-coding genes and contained protein-coding genes mapped from GWS loci such as FURIN, FES, and DRD2. These multi-ancestry pathway results, while more difficult to interpret, may be more generalizable to the global population.
Drug target enrichment results suggest that SA risk is associated with the targets of antipsychotic and antidepressant drug classes. One explanation may be that psychiatric symptoms associated with SA risk are also associated with these drug targets, although the direction of any association of drugs with risk cannot be assumed and was not directly tested here. The SMR analysis of EUR results implicated FES and TIAF1 in SA. FES has previously been implicated in cross-ancestry schizophrenia (53).
Genetic correlations of SA with ADHD, smoking, pain, and risk tolerance remained significant after conditioning SA on both MDD and PTSD, while those for schizophrenia, bipolar disorder, and neuroticism did not. This suggests a potential role for health factors in SA risk that are both shared with and distinct from psychiatric disorders, as proposed in Mann and Rizk’s stress-diathesis model (55) of suicidal behavior based on clinical and biological studies. The suicide diathesis includes altered decision making that may be more pronounced in the context of ADHD and smoking, and may be aggravated by sleep problems. Pain is associated with the stress domain of suicidal behavior, and is also associated with increased access to prescription opioids. Overall, this study leverages genetic data to examine important risk phenotypes that may or may not be present in medical records.
Some limitations of this study should be considered. First, a meta-analysis of such a large number of diverse cohorts, with different assessments of SA, could reduce statistical power by increasing heterogeneity. Our analyses remain still more conservative with the inclusion of age and sex covariates in three of the ISGC cohorts and MVP. However, GWASs of the primary data sets typically produced significant—and high—genetic correlation estimates. GWS loci produced similar effect sizes across cohorts and across fixed and meta-regression models (correlations of EUR and multi-ancestry GWS effect sizes across models exceeded 0.99). Indeed, the apparent consistency of genetic architecture across EUR ISGC and MVP cohorts is important given marked demographic and ascertainment differences.
This study also provides GWAS meta-analyses specific to African and East Asian ancestry admixtures. The lack of GWS loci specific to these SA meta-analyses underscores a strong need for greater ancestral diversity and representation in suicide genetics research. With high variability of sample sizes of individual ISGC and MVP ancestral cohorts (case Ns ranging from 115 to 9,196), some GWASs yielded h2 and rG estimates, while others did not. Variability in rG indicates that increasing the examination of non-European ancestry admixtures in the future will significantly increase the generalizability of the genetic risk signals identified from studies of suicide phenotypes and the portability of polygenic scores. Importantly, broader ancestral representation, particularly from population-dense areas such as India, West Asia, and the Global South, will be critical for improving the rigor and generalizability of GWAS results in future research.
Implicated genes and established genetic relationships with ADHD, smoking, and risk tolerance help to inform our understanding of biological contributions to risk of SA. From a clinical standpoint, impulsivity, smoking status, and risk-taking behaviors are intuitive comorbid indicators of suicide risk. Genetic causal proportion analyses implicate these and other health factors—pulmonary and cardiovascular—in risk for SA. And our preliminary comparison of genetic correlations across SA versus suicide death GWAS cohorts appears to implicate risk tolerance in the severity of the suicide phenotype. Further study comparing suicide death and SA with individuals with suicidal ideation will allow for a comparison of those who think about suicide and those who act. Importantly, genetic risk for SA, calculated in new independent cohorts using these GWAS summary data, will contribute to a deeper understanding of the clinical implications of genetic risk for suicide. The future addition of multiple ancestral cohorts is likely to yield continued discovery and increased opportunity for clinical translation.
1. : Suicide. 2021. https://www.who.int/news-room/fact-sheets/detail/suicide Google Scholar
2. : Preventing Suicide: A Global Imperative. Geneva, World Health Organization, 2014. https://apps.who.int/iris/rest/bitstreams/585331/retrieve Google Scholar
3. : National Survey on Drug Use and Health, 2021. https://www.samhsa.gov/data/release/2021-national-survey-drug-use-and-health-nsduh-releases Google Scholar
4. : CDC WONDER Online Database. 2023. https://wonder.cdc.gov/ Google Scholar
5. : Risk factors for suicidal thoughts and behaviors: a meta-analysis of 50 years of research. Psychol Bull 2017; 143:187–232Crossref, Medline, Google Scholar
6. : Genetics of suicide: a systematic review of twin studies. Wien Klin Wochenschr 2007; 119:463–475Crossref, Medline, Google Scholar
7. : Dissecting the shared genetic architecture of suicide attempt, psychiatric disorders, and known risk factors. Biol Psychiatry 2022; 91:313–327Crossref, Medline, Google Scholar
8. : A genome-wide association study of suicide attempts in the Million Veterans Program identifies evidence of pan-ancestry and ancestry-specific risk loci. Mol Psychiatry 2022; 27:2264–2272Crossref, Medline, Google Scholar
9. : Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J Clin Epidemiol 2016; 70:214–223Crossref, Medline, Google Scholar
10. : Psychiatric genomics: an update and an agenda. Am J Psychiatry 2018; 175:15–27Link, Google Scholar
11. : Human demographic history impacts genetic risk prediction across diverse populations. Am J Hum Genet 2017; 100:635–649Crossref, Medline, Google Scholar
12. : Genome-wide association study of suicide death and polygenic prediction of clinical antecedents. Am J Psychiatry 2020; 177:917–927Link, Google Scholar
13. : VA Suicide Prevention Applications Network: a national health care system-based suicide event tracking system. Public Health Rep 2016; 131:816–821Crossref, Medline, Google Scholar
14. : LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015; 47:291–295Crossref, Medline, Google Scholar
15. : Harmonizing genetic ancestry and self-identified race/ethnicity in genome-wide association studies. Am J Hum Genet 2019; 105:763–772Crossref, Medline, Google Scholar
16. : Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015; 4:7Crossref, Medline, Google Scholar
17. : METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26:2190–2191Crossref, Medline, Google Scholar
18. : Estimating heritability and its enrichment in tissue-specific gene sets in admixed populations. Hum Mol Genet 2021; 30:1521–1534Medline, Google Scholar
19. : The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020; 581:434–443Crossref, Medline, Google Scholar
20. : Cross-national prevalence and risk factors for suicidal ideation, plans and attempts. Br J Psychiatry 2008; 192:98–105Crossref, Medline, Google Scholar
21. : Transethnic genetic-correlation estimates from summary statistics. Am J Hum Genet 2016; 99:76–88Crossref, Medline, Google Scholar
22. : Genetic aetiology of self-harm ideation and behaviour. Sci Rep 2020; 10:9713Crossref, Medline, Google Scholar
23. : Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet 2019; 51:63–75Crossref, Medline, Google Scholar
24. : Identification of common genetic risk variants for autism spectrum disorder. Nat Genet 2019; 51:431–444Crossref, Medline, Google Scholar
25. : Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet 2019; 51:245–257Crossref, Medline, Google Scholar
26. : Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat Genet 2018; 50:920–927Crossref, Medline, Google Scholar
27. : International meta-analysis of PTSD genome-wide association studies identifies sex- and ancestry-specific genetic risk loci. Nat Commun 2019; 10:4558Crossref, Medline, Google Scholar
28. : Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet 2019; 51:793–803Crossref, Medline, Google Scholar
29. : Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum Mol Genet 2018; 27:3641–3649Crossref, Medline, Google Scholar
30. : Genome-wide association study of multisite chronic pain in UK Biobank. PLoS Genet 2019; 15:e1008164Crossref, Medline, Google Scholar
31. : Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet 2019; 51:237–244Crossref, Medline, Google Scholar
32. : Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci 2019; 22:343–352Crossref, Medline, Google Scholar
33. : Genome-wide analysis of insomnia in 1,331,010 individuals identifies new risk loci and functional pathways. Nat Genet 2019; 51:394–403Crossref, Medline, Google Scholar
34. : LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 2017; 33:272–279Crossref, Medline, Google Scholar
35. : Major physical health conditions and risk of suicide. Am J Prev Med 2017; 53:308–315Crossref, Medline, Google Scholar
36. : Suicide. N Engl J Med 2020; 382:266–274Crossref, Medline, Google Scholar
37. : Genomics of body fat percentage may contribute to sex bias in anorexia nervosa. Am J Med Genet B Neuropsychiatr Genet 2019; 180:428–438Crossref, Medline, Google Scholar
38. : Assessment and visualization of phenome-wide causal relationships using genetic data: an application to dental caries and periodontitis. Eur J Hum Genet 2021; 29:300–308Crossref, Medline, Google Scholar
39. : Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun 2018; 9:224Crossref, Medline, Google Scholar
40. : GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 2011; 88:76–82Crossref, Medline, Google Scholar
41. : A global reference for human genetic variation. Nature 2015; 526:68–74Crossref, Medline, Google Scholar
42. : MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 2015; 11:e1004219Crossref, Medline, Google Scholar
43. : Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017; 8:1826Crossref, Medline, Google Scholar
44. : Genetic effects on gene expression across human tissues. Nature 2017; 550:204–213Crossref, Medline, Google Scholar
45. : Drug enrichment and discovery from schizophrenia genome-wide association results: an analysis and visualisation approach. Sci Rep 2017; 7:12460Crossref, Medline, Google Scholar
46. : DGIdb 2.0: mining clinically relevant drug-gene interactions. Nucleic Acids Res 2016; 44:D1036–D1044Crossref, Medline, Google Scholar
47. : The multiplicity of serotonin receptors: uselessly diverse molecules or an embarrassment of riches? Neuroscientist 2000; 6:252–262 Crossref, Google Scholar
48. : Integrative analysis of omics summary data reveals putative mechanisms underlying complex traits. Nat Commun 2018; 9:918Crossref, Medline, Google Scholar
49. : Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet 2016; 48:481–487Crossref, Medline, Google Scholar
50. : Brain expression quantitative trait locus and network analyses reveal downstream effects and putative drivers for brain-related diseases. Nat Genet 2023; 55:377–388Crossref, Medline, Google Scholar
51. : Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun 2019; 10:1776Crossref, Medline, Google Scholar
52. : The NHGRI-EBI GWAS Catalog: knowledgebase and deposition resource. Nucleic Acids Res 2023; 51:D977–D985Crossref, Medline, Google Scholar
53. : Genome-wide association study detected novel susceptibility genes for schizophrenia and shared trans-populations/diseases genetic effect. Schizophr Bull 2019; 45:824–834Crossref, Medline, Google Scholar
54. : Hypothalamus-pituitary-adrenal system regulation and suicidal behavior in depression. Biol Psychiatry 2005; 57:336–342Crossref, Medline, Google Scholar
55. : A brain-centric model of suicidal behavior. Am J Psychiatry 2020; 177:902–916Link, Google Scholar



